| Maintainer: | Mark van der Loo <mark.vanderloo@gmail.com> |
| License: | GPL-3 |
| Title: | Data Validation Infrastructure |
| LazyData: | no |
| Type: | Package |
| LazyLoad: | yes |
| Description: | Declare data validation rules and data quality indicators; confront data with them and analyze or visualize the results. The package supports rules that are per-field, in-record, cross-record or cross-dataset. Rules can be automatically analyzed for rule type and connectivity. Supports checks implied by an SDMX DSD file as well. See also Van der Loo and De Jonge (2018) <doi:10.1002/9781118897126>, Chapter 6 and the JSS paper (2021) <doi:10.18637/jss.v097.i10>. |
| Version: | 1.1.7 |
| Depends: | R (≥ 3.5.0), methods |
| URL: | https://github.com/data-cleaning/validate |
| BugReports: | https://github.com/data-cleaning/validate/issues |
| Imports: | stats, graphics, grid, settings, yaml |
| Suggests: | rsdmx, tinytest (≥ 0.9.6), knitr, bookdown, lumberjack, rmarkdown |
| VignetteBuilder: | knitr |
| Collate: | 'rule.R' 'sugar.R' 'validate_pkg.R' 'parse.R' 'expressionset.R' 'indicator.R' 'validator.R' 'confrontation.R' 'compare.R' 'factory.R' 'genericrules.R' 'lumberjack.R' 'plot.R' 'retailers.R' 'run_validation.R' 'sdmx.R' 'syntax.R' 'utils.R' 'yaml.R' |
| RoxygenNote: | 7.3.2 |
| Encoding: | UTF-8 |
| NeedsCompilation: | yes |
| Packaged: | 2025-12-10 15:28:11 UTC; mark |
| Author: | Mark van der Loo |
| Repository: | CRAN |
| Date/Publication: | 2025-12-10 17:10:02 UTC |
Data Validation Infrastructure
Description
Data often suffer from errors and missing values. A necessary step before data
analysis is verifying and validating your data. Package validate is a
toolbox for creating validation rules and checking data against these rules.
Getting started
The easiest way to get started is through the examples given in check_that.
The general workflow in validate follows the following pattern.
Define a set of rules or quality indicator using
validatororindicator.-
confrontdata with the rules or indicators, Examine the results either graphically or by summary.
There are several convenience functions that allow one to define rules from the commandline, through a (freeform or yaml) file and to investigate and maintain the rules themselves. Please have a look at the cookbook for a comprehensive introduction.
Author(s)
Maintainer: Mark van der Loo mark.vanderloo@gmail.com (ORCID)
Authors:
Edwin de Jonge (ORCID)
Other contributors:
Paul Hsieh [contributor]
References
An overview of this package, its underlying ideas and many examples can be found in MPJ van der Loo and E. de Jonge (2018) Statistical data cleaning with applications in R John Wiley & Sons.
Please use citation("validate") to get a citation for (scientific)
publications.
See Also
Useful links:
Report bugs at https://github.com/data-cleaning/validate/issues
A consistent set membership operator
Description
A set membership operator like %in% that handles
NA more consistently with R's other logical comparison operators.
Usage
x %vin% table
Arguments
x |
vector or |
table |
vector or |
Details
R's basic comparison operators (almost) always return NA when one
of the operands is NA. The %in% operator is an exception.
Compare for example NA %in% NA with NA == NA: the first
results in TRUE, while the latter results in NA as expected.
The %vin% operator acts consistent with operators such as ==.
Specifically, NA results in the following cases.
For each position where
xisNA, the result isNA.When
tablecontains anNA, each non-matched value inxresults inNA.
Examples
# we cannot be sure about the first element:
c(NA, "a") %vin% c("a","b")
# we cannot be sure about the 2nd and 3rd element (but note that they
# cannot both be TRUE):
c("a","b","c") %vin% c("a",NA)
# we can be sure about all elements:
c("a","b") %in% character(0)
Combine two indicator objects
Description
Combine two indicator objects by addition. A new indicator
object is created with default (global) option values. Previously set options
are ignored.
Usage
## S4 method for signature 'indicator,indicator'
e1 + e2
Arguments
e1 |
|
e2 |
Examples
indicator(mean(x)) + indicator(x/median(x))
Combine two validator objects
Description
Combine two validator objects by addition. A new validator
object is created with default (global) option values. Previously set options
are ignored.
Usage
## S4 method for signature 'validator,validator'
e1 + e2
Arguments
e1 |
|
e2 |
Note
The names of the resulting object are made unique using make.names.
See Also
Other validator-methods:
plot,validator-method,
validator
Examples
validator(x>0) + validator(x<=1)
Services for extending 'validate'
Description
Functions exported silently to allow for cross-package inheritance
of the expressionset object. These functions are never
needed in scripts or statistical production code.
Usage
.PKGOPT(..., .__defaults = FALSE, .__reset = FALSE)
.ini_expressionset_cli(obj, ..., .prefix = "R")
.ini_expressionset_df(obj, dat, .prefix = "R")
.ini_expressionset_yml(obj, file, .prefix = "R")
.show_expressionset(obj)
.get_exprs(
x,
...,
expand_assignments = FALSE,
expand_groups = TRUE,
vectorize = TRUE,
replace_dollar = TRUE,
replace_in = TRUE,
lin_eq_eps = x$options("lin.eq.eps"),
lin_ineq_eps = x$options("lin.ineq.eps"),
dat = NULL
)
.blocks_expressionset(x)
Arguments
... |
Comma-separated list of expressions |
.__defaults |
toggle default options |
.__reset |
togle reset options |
obj |
an expressionset object |
.prefix |
Prefix to use in default names. |
dat |
Optionally, a |
file |
a filename |
x |
An expressionset object |
expand_assignments |
Substitute assignments? |
expand_groups |
Expand groups? |
vectorize |
Vectorize if-statements? |
replace_dollar |
Replace dollar with bracket index? |
Details
This function is aimed at developers importing the package and not at direct users of validate.
Replace a subset of an expressionset with another expressionset
Description
Replace a subset of an expressionset with another expressionset
Usage
## S4 replacement method for signature 'expressionset'
x[i] <- value
Arguments
x |
an R object inheriting from |
i |
a |
value |
an R object of the same class as |
Select a subset
Description
Select a subset
Usage
## S4 method for signature 'expressionset'
x[i, j, ..., drop = TRUE]
## S4 method for signature 'expressionset'
x[[i, j, ..., exact = TRUE]]
## S4 method for signature 'confrontation'
x[i, j, ..., drop = TRUE]
Arguments
x |
An R object |
i |
an index (numeric, boolean, character) |
j |
not implemented |
... |
Arguments to be passed to other methods |
drop |
not implemented |
exact |
Not implemented |
Value
An new object, of the same class as x subsetted according to i.
Details
The options attribute will be cloned
See Also
Other confrontation-methods:
as.data.frame,confrontation-method,
confront(),
confrontation-class,
errors(),
event(),
keyset(),
length,expressionset-method,
values()
Replace a rule in a ruleseta
Description
Replace a rule in a ruleseta
Usage
## S4 replacement method for signature 'expressionset'
x[[i]] <- value
Arguments
x |
an R object |
i |
index of length 1 |
value |
object of class |
Add indicator values as columns to a data frame
Description
Compute and add externally defined indicators to data frame. If necessary, values are recycled over records.
Usage
add_indicators(dat, x)
Arguments
dat |
|
x |
|
Value
dat with extra columns defined by x attached.
Examples
ii <- indicator(
hihi = 2*sqrt(height)
, haha = log10(weight)
, lulz = mean(height)
, wo0t = median(weight)
)
# note: mean and median are repeated
add_indicators(women, ii)
# compute indicators first, then add
out <- confront(women, ii)
add_indicators(women, out)
Aggregate validation results
Description
Aggregate results of a validation.
Usage
## S4 method for signature 'validation'
aggregate(x, by = c("rule", "record"), drop = TRUE, ...)
Arguments
x |
An object of class |
by |
Report on violations per rule (default) or per record? |
drop |
drop list attribute if the result is list of length 1 |
... |
Arguments to be passed to or from other methods. |
Value
By default, a data.frame with the following columns.
| keys | If confront was called with key= |
npass | Number of items passed |
nfail | Number of items failing |
nNA | Number of items resulting in NA |
rel.pass | Relative number of items passed |
rel.fail | Relative number of items failing |
rel.NA | Relative number of items resulting in NA
|
If by='rule' the relative numbers are computed with respect to the number
of records for which the rule was evaluated. If by='record' the relative numbers
are computed with respect to the number of rules the record was tested agains.
When by='record' and not all validation results have the same dimension structure,
a list of data.frames is returned.
See Also
Other validation-methods:
all,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
compare(),
confront(),
event(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
summary(),
validation-class,
values()
Examples
data(retailers)
retailers$id <- paste0("ret",1:nrow(retailers))
v <- validator(
staff.costs/staff < 25
, turnover + other.rev==total.rev)
cf <- confront(retailers,v,key="id")
a <- aggregate(cf,by='record')
head(a)
# or, get a sorted result:
s <- sort(cf, by='record')
head(s)
Test if all validations resulted in TRUE
Description
Test if all validations resulted in TRUE
Usage
## S4 method for signature 'validation'
all(x, ..., na.rm = FALSE)
Arguments
x |
|
... |
ignored |
na.rm |
[ |
See Also
Other validation-methods:
aggregate,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
compare(),
confront(),
event(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
summary(),
validation-class,
values()
Examples
val <- check_that(women, height>60, weight>0)
all(val)
Test if any validation resulted in TRUE
Description
Test if any validation resulted in TRUE
Usage
## S4 method for signature 'validation'
any(x, ..., na.rm = FALSE)
Arguments
x |
|
... |
ignored |
na.rm |
[ |
See Also
Other validation-methods:
aggregate,validation-method,
all,validation-method,
barplot,validation-method,
check_that(),
compare(),
confront(),
event(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
summary(),
validation-class,
values()
Examples
val <- check_that(women, height>60, weight>0)
any(val)
Coerce to data.frame
Description
Coerce to data.frame
Usage
as.data.frame(x, row.names = NULL, optional = FALSE, ...)
Arguments
x |
Object to coerce |
row.names |
ignored |
optional |
ignored |
... |
arguments passed to other methods |
See Also
Other expressionset-methods:
as.data.frame,expressionset-method,
created(),
description(),
label(),
meta(),
names<-,rule,character-method,
origin(),
plot,validator-method,
summary(),
variables(),
voptions()
Translate cellComparison objects to data frame
Description
Versions of a data set can be cellwise compared using
cells. The result is a cellComparison object,
which can usefully be translated into a data frame.
Usage
## S4 method for signature 'cellComparison'
as.data.frame(x, row.names = NULL, optional = FALSE, ...)
Arguments
x |
Object to coerce |
row.names |
ignored |
optional |
ignored |
... |
arguments passed to other methods |
Value
A data frame with the following columns.
status: Row names of thecellComparisonobject.version: Column names of thecellComparisonobject.count: Contents of thecellComparisonobject.
See Also
Other comparing:
as.data.frame,validatorComparison-method,
barplot,cellComparison-method,
barplot,validatorComparison-method,
cells(),
compare(),
match_cells(),
plot,cellComparison-method,
plot,validatorComparison-method
Examples
data(retailers)
# start with raw data
step0 <- retailers
# impute turnovers
step1 <- step0
step1$turnover[is.na(step1$turnover)] <- mean(step1$turnover,na.rm=TRUE)
# flip sign of negative revenues
step2 <- step1
step2$other.rev <- abs(step2$other.rev)
# create an overview of differences, comparing to the previous step
cells(raw = step0, imputed = step1, flipped = step2, compare="sequential")
# create an overview of differences compared to raw data
out <- cells(raw = step0, imputed = step1, flipped = step2)
out
# Graphical overview of the changes
plot(out)
barplot(out)
# transform data to data.frame (easy for use with ggplot)
as.data.frame(out)
Coerce a confrontation object to data frame
Description
Results of confronting data with validation rules or indicators
are created by a confrontation. The result is an
object (inheriting from) confrontation.
Usage
## S4 method for signature 'confrontation'
as.data.frame(x, row.names = NULL, optional = FALSE, ...)
Arguments
x |
Object to coerce |
row.names |
ignored |
optional |
ignored |
... |
arguments passed to other methods |
Value
A data.frame with columns
keyWhere relevant, and only ifkeywas specified in the call toconfrontnameName of the rulevalueValue after evaluationexpressionevaluated expression
See Also
Other confrontation-methods:
[,expressionset-method,
confront(),
confrontation-class,
errors(),
event(),
keyset(),
length,expressionset-method,
values()
Examples
cf <- check_that(women, height > 0, sd(weight) > 0)
as.data.frame(cf)
# add id-column
women$id <- letters[1:15]
i <- indicator(mw = mean(weight), ratio = weight/height)
as.data.frame(confront(women, i, key="id"))
Translate an expressionset to data.frame
Description
Expressions are deparsed and combined in a data.frame with (some
of) their metadata. Observe that some information may be lost (e.g. options
local to the object).
Usage
## S4 method for signature 'expressionset'
as.data.frame(x, expand_assignments = TRUE, ...)
Arguments
x |
Object to coerce |
expand_assignments |
Toggle substitution of ':=' assignments. |
... |
arguments passed to other methods |
Value
A data.frame with elements rule, name,
label, origin, description, and created.
See Also
Other expressionset-methods:
as.data.frame(),
created(),
description(),
label(),
meta(),
names<-,rule,character-method,
origin(),
plot,validator-method,
summary(),
variables(),
voptions()
Translate a validatorComparison object to data frame
Description
The performance of versions of a data set with regard to rule-based quality
requirements can be compared using using compare. The result is a
validatorComparison object, which can usefully be translated into a data
frame.
Usage
## S4 method for signature 'validatorComparison'
as.data.frame(x, row.names = NULL, optional = FALSE, ...)
Arguments
x |
Object to coerce |
row.names |
ignored |
optional |
ignored |
... |
arguments passed to other methods |
Value
A data frame with the following columns.
status: Row names of thevalidatorComparisonobject.version: Column names of thevalidatorComparisonobject.count: Contents of thevalidatorComparisonobject.
See Also
Other comparing:
as.data.frame,cellComparison-method,
barplot,cellComparison-method,
barplot,validatorComparison-method,
cells(),
compare(),
match_cells(),
plot,cellComparison-method,
plot,validatorComparison-method
Examples
data(retailers)
rules <- validator(turnover >=0, staff>=0, other.rev>=0)
# start with raw data
step0 <- retailers
# impute turnovers
step1 <- step0
step1$turnover[is.na(step1$turnover)] <- mean(step1$turnover,na.rm=TRUE)
# flip sign of negative revenues
step2 <- step1
step2$other.rev <- abs(step2$other.rev)
# create an overview of differences, comparing to the previous step
compare(rules, raw = step0, imputed = step1, flipped = step2, how="sequential")
# create an overview of differences compared to raw data
out <- compare(rules, raw = step0, imputed = step1, flipped = step2)
out
# graphical overview
plot(out)
barplot(out)
# transform data to data.frame (easy for use with ggplot)
as.data.frame(out)
Barplot of cellComparison object
Description
Versions of a data set can be compared cell by cell using cells.
The result is a cellComparison object. This method creates a stacked bar
plot of the results. See also plot,cellComparison-method for a
line chart.
Usage
## S4 method for signature 'cellComparison'
barplot(
height,
las = 1,
cex.axis = 0.8,
cex.legend = cex.axis,
wrap = TRUE,
...
)
Arguments
height |
object of class |
las |
[ |
cex.axis |
[ |
cex.legend |
[ |
wrap |
[ |
... |
Graphical parameters passed to |
Note
Before plotting, underscores (_) and dots (.) in x-axis
labels are replaced with spaces.
See Also
Other comparing:
as.data.frame,cellComparison-method,
as.data.frame,validatorComparison-method,
barplot,validatorComparison-method,
cells(),
compare(),
match_cells(),
plot,cellComparison-method,
plot,validatorComparison-method
Plot number of violations
Description
Plot number of violations
Usage
## S4 method for signature 'validation'
barplot(
height,
...,
order_by = c("fails", "passes", "nNA"),
stack_by = c("fails", "passes", "nNA"),
topn = Inf,
add_legend = TRUE,
add_exprs = TRUE,
colors = c(fails = "#FB9A99", passes = "#B2DF8A", nNA = "#FDBF6F")
)
Arguments
height |
an R object defining height of bars (here, a |
... |
parameters to be passed to |
order_by |
(single |
stack_by |
(3-vector of |
topn |
If specified, plot only the top n most violated calls |
add_legend |
Display legend? |
add_exprs |
Display rules? |
colors |
Bar colors for validations yielding NA or a violation |
Value
A list, containing the bar locations as in barplot
Credits
The default colors were generated with the RColorBrewer package of Erich Neuwirth.
See Also
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
check_that(),
compare(),
confront(),
event(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
summary(),
validation-class,
values()
Examples
data(retailers)
cf <- check_that(retailers
, staff.costs < total.costs
, turnover + other.rev == total.rev
, other.rev > 0
, total.rev > 0)
barplot(cf)
Barplot of validatorComparison object
Description
The performance of versions of a data set with regard to rule-based quality
requirements can be compared using using compare. The result is a
validatorComparison object. This method creates a stacked bar plot of
the results. See also plot,validatorComparison-method for a line
chart.
Usage
## S4 method for signature 'validatorComparison'
barplot(
height,
las = 1,
cex.axis = 0.8,
cex.legend = cex.axis,
wrap = TRUE,
...
)
Arguments
height |
object of class |
las |
[ |
cex.axis |
[ |
cex.legend |
[ |
wrap |
[ |
... |
Graphical parameters passed to |
Note
Before plotting, underscores (_) and dots (.) in x-axis labels
are replaced with spaces.
See Also
Other comparing:
as.data.frame,cellComparison-method,
as.data.frame,validatorComparison-method,
barplot,cellComparison-method,
cells(),
compare(),
match_cells(),
plot,cellComparison-method,
plot,validatorComparison-method
Examples
data(retailers)
rules <- validator(turnover >=0, staff>=0, other.rev>=0)
# start with raw data
step0 <- retailers
# impute turnovers
step1 <- step0
step1$turnover[is.na(step1$turnover)] <- mean(step1$turnover,na.rm=TRUE)
# flip sign of negative revenues
step2 <- step1
step2$other.rev <- abs(step2$other.rev)
# create an overview of differences, comparing to the previous step
compare(rules, raw = step0, imputed = step1, flipped = step2, how="sequential")
# create an overview of differences compared to raw data
out <- compare(rules, raw = step0, imputed = step1, flipped = step2)
out
# graphical overview
plot(out)
barplot(out)
# transform data to data.frame (easy for use with ggplot)
as.data.frame(out)
Cell counts and differences for a series of datasets
Description
Cell counts and differences for a series of datasets
Usage
cells(..., .list = NULL, compare = c("to_first", "sequential"))
Arguments
... |
For |
.list |
A |
compare |
How to compare the datasets. |
Value
An object of class cellComparison, which is really an array
with a few extra attributes. It counts the total number of cells, the number of
missings, the number of altered values and changes therein as compared to
the reference defined in how.
Comparing datasets cell by cell
When comparing the contents of two data sets, the total number of cells in the current data set can be partitioned as in the following figure.
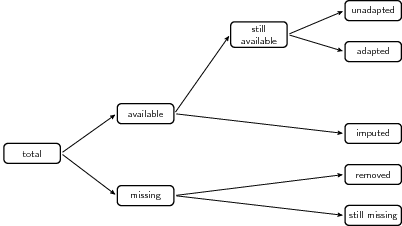
This function computes the partition for two or more
datasets, comparing the current set to the first (default) or to the
previous (by setting compare='sequential').
Details
This function assumes that the datasets have the same dimensions and that both rows and columns are ordered similarly.
References
The figure is reproduced from MPJ van der Loo and E. De Jonge (2018) Statistical Data Cleaning with applications in R (John Wiley & Sons).
See Also
Other comparing:
as.data.frame,cellComparison-method,
as.data.frame,validatorComparison-method,
barplot,cellComparison-method,
barplot,validatorComparison-method,
compare(),
match_cells(),
plot,cellComparison-method,
plot,validatorComparison-method
Examples
data(retailers)
# start with raw data
step0 <- retailers
# impute turnovers
step1 <- step0
step1$turnover[is.na(step1$turnover)] <- mean(step1$turnover,na.rm=TRUE)
# flip sign of negative revenues
step2 <- step1
step2$other.rev <- abs(step2$other.rev)
# create an overview of differences, comparing to the previous step
cells(raw = step0, imputed = step1, flipped = step2, compare="sequential")
# create an overview of differences compared to raw data
out <- cells(raw = step0, imputed = step1, flipped = step2)
out
# Graphical overview of the changes
plot(out)
barplot(out)
# transform data to data.frame (easy for use with ggplot)
as.data.frame(out)
Simple data validation interface
Description
Simple data validation interface
Usage
check_that(dat, ...)
Arguments
dat |
an R object carrying data |
... |
a comma-separated set of validating expressions. |
Value
An object of class validation
Details
Creates an object of class validator and confronts it with the data.
This function is easy to use in combination with the magrittr pipe operator.
See Also
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
barplot,validation-method,
compare(),
confront(),
event(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
summary(),
validation-class,
values()
Examples
cf <- check_that(women, height>0, height/weight < 0.5)
cf
summary(cf)
barplot(cf)
## Not run:
# this works only after loading the 'magrittr' package
women %>%
check_that(height>0, height/weight < 0.5) %>%
summary()
## End(Not run)
Compare similar data sets
Description
Compare versions of a data set by comparing their performance against a
set of rules or other quality indicators. This function takes two or
more data sets and compares the perfomance of data set 2,3,\ldots
against that of the first data set (default) or to the previous one
(by setting how='sequential').
Usage
compare(x, ...)
## S4 method for signature 'validator'
compare(x, ..., .list = list(), how = c("to_first", "sequential"))
## S4 method for signature 'indicator'
compare(x, ..., .list = NULL)
Arguments
x |
An R object |
... |
data frames, comma separated. Names become column names in the output. |
.list |
Optional list of data sets, will be concatenated with |
how |
how to compare |
Value
For validator: An array where each column represents
one dataset.
The rows count the following attributes:
Number of validations performed
Number of validations that evaluate to
NA(unverifiable)Number of validations that evaluate to a logical (verifiable)
Number of validations that evaluate to
TRUENumber of validations that evaluate to
FALSENumber of extra validations that evaluate to
NA(new unverifiable)Number of validations that still evaluate to
NA(still unverifialble)Number of validations that still evaluate to
TRUENumber of extra validations that evaluate to
TRUENumber of validations that still evaluate to
FALSENumber of extra validations that evaluate to
FALSE
For indicator: A list with the following components:
numeric: An array collecting results of scalar indicator (e.g.mean(x)).nonnumeric: An array collecting results of nonnumeric scalar indicators (e.g. names(which.max(table(x))))array: A list of arrays, collecting results of vector-indicators (e.g. x/mean(x))
Comparing datasets by performance against validator objects
Suppose we have a current and a previous version of a data set. Both
can be inspected by confronting them with a rule set.
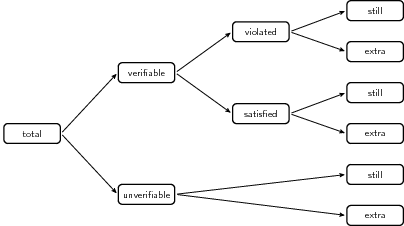
The status changes in rule violations can be partitioned as shown in the
following figure.
 This function computes the partition for two or more
datasets, comparing the current set to the first (default) or to the
previous (by setting
This function computes the partition for two or more
datasets, comparing the current set to the first (default) or to the
previous (by setting compare='sequential').
References
The figure is reproduced from MPJ van der Loo and E. De Jonge (2018) Statistical Data Cleaning with applications in R (John Wiley & Sons).
See Also
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
confront(),
event(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
summary(),
validation-class,
values()
Other comparing:
as.data.frame,cellComparison-method,
as.data.frame,validatorComparison-method,
barplot,cellComparison-method,
barplot,validatorComparison-method,
cells(),
match_cells(),
plot,cellComparison-method,
plot,validatorComparison-method
Examples
data(retailers)
rules <- validator(turnover >=0, staff>=0, other.rev>=0)
# start with raw data
step0 <- retailers
# impute turnovers
step1 <- step0
step1$turnover[is.na(step1$turnover)] <- mean(step1$turnover,na.rm=TRUE)
# flip sign of negative revenues
step2 <- step1
step2$other.rev <- abs(step2$other.rev)
# create an overview of differences, comparing to the previous step
compare(rules, raw = step0, imputed = step1, flipped = step2, how="sequential")
# create an overview of differences compared to raw data
out <- compare(rules, raw = step0, imputed = step1, flipped = step2)
out
# graphical overview
plot(out)
barplot(out)
# transform data to data.frame (easy for use with ggplot)
as.data.frame(out)
Confront data with a (set of) expressionset(s)
Description
An expressionset is a general class storing rich expressions (basically
expressions and some meta data) which we call 'rules'. Examples of
expressionset implementations are validator objects, storing
validation rules and indicator objects, storing data quality
indicators. The confront function evaluates the expressions one by one
on a dataset while recording some process meta data. All results are stored in
a (subclass of a) confrontation object.
Usage
confront(dat, x, ref, ...)
## S4 method for signature 'data.frame,indicator,ANY'
confront(dat, x, key = NULL, ...)
## S4 method for signature 'data.frame,indicator,environment'
confront(dat, x, ref, key = NULL, ...)
## S4 method for signature 'data.frame,indicator,data.frame'
confront(dat, x, ref, key = NULL, ...)
## S4 method for signature 'data.frame,indicator,list'
confront(dat, x, ref, key = NULL, ...)
## S4 method for signature 'data.frame,validator,ANY'
confront(dat, x, key = NULL, ...)
## S4 method for signature 'data.frame,validator,environment'
confront(dat, x, ref, key = NULL, ...)
## S4 method for signature 'data.frame,validator,data.frame'
confront(dat, x, ref, key = NULL, ...)
## S4 method for signature 'data.frame,validator,list'
confront(dat, x, ref, key = NULL, ...)
Arguments
dat |
An R object carrying data |
x |
An R object carrying |
ref |
Optionally, an R object carrying reference data. See examples for usage. |
... |
Options used at execution time (especially |
key |
(optional) name of identifying variable in x. |
Reference data
Reference data is typically a list with a items such as
a code list, or a data frame of which rows match the rows of the
data under scrutiny.
See Also
Other confrontation-methods:
[,expressionset-method,
as.data.frame,confrontation-method,
confrontation-class,
errors(),
event(),
keyset(),
length,expressionset-method,
values()
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
compare(),
event(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
summary(),
validation-class,
values()
Other indication-methods:
event(),
indication-class,
summary()
Examples
# a basic validation example
v <- validator(height/weight < 0.5, mean(height) >= 0)
cf <- confront(women, v)
summary(cf)
plot(cf)
as.data.frame(cf)
# an example checking metadata
v <- validator(nrow(.) == 15, ncol(.) > 2)
summary(confront(women, v))
# An example using reference data
v <- validator(weight == ref$weight)
summary(confront(women, v, women))
# Usging custom names for reference data
v <- validator(weight == test$weight)
summary( confront(women,v, list(test=women)) )
# Reference data in an environment
e <- new.env()
e$test <- women
v <- validator(weight == test$weight)
summary( confront(women, v, e) )
# the effect of using a key
w <- women
w$id <- letters[1:nrow(w)]
v <- validator(weight == ref$weight)
# with complete data; already matching
values( confront(w, v, w, key='id'))
# with scrambled rows in reference data (reference gets sorted according to dat)
i <- sample(nrow(w))
values(confront(w, v, w[i,],key='id'))
# with incomplete reference data
values(confront(w, v, w[1:10,],key='id'))
Superclass storing results of confronting data with rules
Description
Superclass storing results of confronting data with rules
Details
This class is aimed at developers of this package or packages depending on
it. It is the parent of classes indication and
validation which are user-facing.
Using confront, a set of rules can be executed in the context
of one or more (nested) environments holding data. The results of such evaluations
are stored in a confrontation object along with metadata.
We strongly advise against accessing the data fields or methods internal to this object directly, as we may change or remove them without notice. Use the exported methods listed below in stead.
See Also
Other confrontation-methods:
[,expressionset-method,
as.data.frame,confrontation-method,
confront(),
errors(),
event(),
keyset(),
length,expressionset-method,
values()
Check records using a predifined table of (im)possible values
Description
Given a set of keys or key combinations, check whether all thos combinations occur, or check that they do not occur. Supports globbing and regular expressions.
Usage
contains_exactly(keys, by = NULL, allow_duplicates = FALSE)
contains_at_least(keys, by = NULL)
contains_at_most(keys, by = NULL)
does_not_contain(keys)
Arguments
keys |
A data frame or bare (unquoted) name of a data
frame passed as a reference to |
by |
A bare (unquoted) variable or list of variable names that occur in the data under scrutiny. The data will be split into groups according to these variables and the check is performed on each group. |
allow_duplicates |
|
Details
contains_exactly | dataset contains exactly the key set, no more, no less. |
contains_at_least | dataset contains at least the given keys. |
contains_at_most | all keys in the data set are contained the given keys. |
does_not_contain | The keys are interpreted as forbidden key combinations. |
Value
For contains_exactly, contains_at_least, and
contains_at_most a logical vector with one entry for each
record in the dataset. Any group not conforming to the test keys will have
FALSE assigned to each record in the group (see examples).
For contains_at_least: a logical vector equal to the number of
records under scrutiny. It is FALSE where key combinations do not match
any value in keys.
For does_not_contain: a logical vector with size equal to the
number of records under scrutiny. It is FALSE where key combinations
do not match any value in keys.
Globbing
Globbing is a simple method of defining string patterns where the asterisks
(*) is used a wildcard. For example, the globbing pattern
"abc*" stands for any string starting with "abc".
See Also
Other cross-record-helpers:
do_by(),
exists_any(),
hb(),
hierarchy(),
is_complete(),
is_linear_sequence(),
is_unique()
Examples
## Check that data is present for all quarters in 2018-2019
dat <- data.frame(
year = rep(c("2018","2019"),each=4)
, quarter = rep(sprintf("Q%d",1:4), 2)
, value = sample(20:50,8)
)
# Method 1: creating a data frame in-place (only for simple cases)
rule <- validator(contains_exactly(
expand.grid(year=c("2018","2019"), quarter=c("Q1","Q2","Q3","Q4"))
)
)
out <- confront(dat, rule)
values(out)
# Method 2: pass the keyset to 'confront', and reference it in the rule.
# this scales to larger key sets but it needs a 'contract' between the
# rule definition and how 'confront' is called.
keyset <- expand.grid(year=c("2018","2019"), quarter=c("Q1","Q2","Q3","Q4"))
rule <- validator(contains_exactly(all_keys))
out <- confront(dat, rule, ref=list(all_keys = keyset))
values(out)
## Globbing (use * as a wildcard)
# transaction data
transactions <- data.frame(
sender = c("S21", "X34", "S45","Z22")
, receiver = c("FG0", "FG2", "DF1","KK2")
, value = sample(70:100,4)
)
# forbidden combinations: if the sender starts with "S",
# the receiver can not start "FG"
forbidden <- data.frame(sender="S*",receiver = "FG*")
rule <- validator(does_not_contain(glob(forbidden_keys)))
out <- confront(transactions, rule, ref=list(forbidden_keys=forbidden))
values(out)
## Quick interactive testing
# use 'with':
with(transactions, does_not_contain(forbidden))
## Grouping
# data in 'long' format
dat <- expand.grid(
year = c("2018","2019")
, quarter = c("Q1","Q2","Q3","Q4")
, variable = c("import","export")
)
dat$value <- sample(50:100,nrow(dat))
periods <- expand.grid(
year = c("2018","2019")
, quarter = c("Q1","Q2","Q3","Q4")
)
rule <- validator(contains_exactly(all_periods, by=variable))
out <- confront(dat, rule, ref=list(all_periods=periods))
values(out)
# remove one export record
dat1 <- dat[-15,]
out1 <- confront(dat1, rule, ref=list(all_periods=periods))
values(out1)
values(out1)
Creation timestamp
Description
Creation timestamp
Usage
created(x, ...)
created(x) <- value
## S4 method for signature 'rule'
created(x, ...)
## S4 replacement method for signature 'rule,POSIXct'
created(x) <- value
## S4 method for signature 'expressionset'
created(x, ...)
## S4 replacement method for signature 'expressionset,POSIXct'
created(x) <- value
Arguments
x |
and R object |
... |
Arguments to be passed to other methods |
value |
Value to set |
Value
A POSIXct vector.
See Also
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
description(),
label(),
meta(),
names<-,rule,character-method,
origin(),
plot,validator-method,
summary(),
variables(),
voptions()
Examples
# retrieve properties
v <- validator(turnover > 0, staff.costs>0)
# number of rules in v:
length(v)
# per-rule
created(v)
origin(v)
names(v)
# set properties
names(v)[1] <- "p1"
label(v)[1] <- "turnover positive"
description(v)[1] <- "
According to the official definition,
only positive values can be considered
valid turnovers.
"
# short description is also printed:
v
# print all info for first rule
v[[1]]
# retrieve properties
v <- validator(turnover > 0, staff.costs>0)
# number of rules in v:
length(v)
# per-rule
created(v)
origin(v)
names(v)
# set properties
names(v)[1] <- "p1"
label(v)[1] <- "turnover positive"
description(v)[1] <- "
According to the official definition,
only positive values can be considered
valid turnovers.
"
# short description is also printed:
v
# print all info for first rule
v[[1]]
Rule description
Description
A longer (typically one-paragraph) description of a rule.
Usage
description(x, ...)
description(x) <- value
## S4 method for signature 'rule'
description(x, ...)
## S4 replacement method for signature 'rule,character'
description(x) <- value
## S4 method for signature 'expressionset'
description(x, ...)
## S4 replacement method for signature 'expressionset,character'
description(x) <- value
Arguments
x |
and R object |
... |
Arguments to be passed to other methods |
value |
Value to set |
Value
A character vector.
See Also
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
label(),
meta(),
names<-,rule,character-method,
origin(),
plot,validator-method,
summary(),
variables(),
voptions()
Examples
# retrieve properties
v <- validator(turnover > 0, staff.costs>0)
# number of rules in v:
length(v)
# per-rule
created(v)
origin(v)
names(v)
# set properties
names(v)[1] <- "p1"
label(v)[1] <- "turnover positive"
description(v)[1] <- "
According to the official definition,
only positive values can be considered
valid turnovers.
"
# short description is also printed:
v
# print all info for first rule
v[[1]]
# retrieve properties
v <- validator(turnover > 0, staff.costs>0)
# number of rules in v:
length(v)
# per-rule
created(v)
origin(v)
names(v)
# set properties
names(v)[1] <- "p1"
label(v)[1] <- "turnover positive"
description(v)[1] <- "
According to the official definition,
only positive values can be considered
valid turnovers.
"
# short description is also printed:
v
# print all info for first rule
v[[1]]
split-apply-combine for vectors, with equal-length outptu
Description
Group x by one or more categorical variables, compute
an aggregate, repeat that aggregate to match the size of the
group, and combine results. The functions sum_by and
so on are convenience wrappers that call do_by internally.
Usage
do_by(x, by, fun, ...)
sum_by(x, by, na.rm = FALSE)
mean_by(x, by, na.rm = FALSE)
min_by(x, by, na.rm = FALSE)
max_by(x, by, na.rm = FALSE)
Arguments
x |
A bare variable name |
by |
a bare variable name, or a list of bare variable names, used to
split |
fun |
|
... |
passed as extra arguments to |
na.rm |
Toggle ignoring |
See Also
Other cross-record-helpers:
contains_exactly(),
exists_any(),
hb(),
hierarchy(),
is_complete(),
is_linear_sequence(),
is_unique()
Examples
x <- 1:10
y <- rep(letters[1:2], 5)
do_by(x, by=y, fun=max)
do_by(x, by=y, fun=sum)
Get messages from a confrontation object
Description
Get messages from a confrontation object
Usage
errors(x, ...)
## S4 method for signature 'confrontation'
errors(x, ...)
## S4 method for signature 'confrontation'
warnings(x, ...)
Arguments
x |
An object of class |
... |
Arguments to be passed to other methods. |
See Also
Other confrontation-methods:
[,expressionset-method,
as.data.frame,confrontation-method,
confront(),
confrontation-class,
event(),
keyset(),
length,expressionset-method,
values()
Examples
# create an error, by using a non-existent variable name
cf <- check_that(women, hite > 0, weight > 0)
# retrieve error messages
errors(cf)
Get or set event information metadata from a 'confrontation' object.
Description
The purpose of event information is to store information that allows for identification of the confronting event.
Usage
event(x)
event(x) <- value
## S4 method for signature 'confrontation'
event(x)
## S4 replacement method for signature 'confrontation'
event(x) <- value
Arguments
x |
an object of class |
value |
|
Value
A a character vector with elements
"agent", which defaults to the R version and platform returned by
R.version, a timestamp ("time") in ISO 8601 format and a
"actor" which is the user name returned by Sys.info(). The
last element is called "trigger" (default NA_character_), which
can be used to administrate the event that triggered the confrontation.
References
Mark van der Loo and Olav ten Bosch (2017) Design of a generic machine-readable validation report structure, version 1.0.0.
See Also
Other confrontation-methods:
[,expressionset-method,
as.data.frame,confrontation-method,
confront(),
confrontation-class,
errors(),
keyset(),
length,expressionset-method,
values()
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
compare(),
confront(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
summary(),
validation-class,
values()
Other indication-methods:
confront(),
indication-class,
summary()
Examples
data(retailers)
rules <- validator(turnover >= 0, staff >=0)
cf <- confront(retailers, rules)
event(cf)
# adapt event information
u <- event(cf)
u["trigger"] <- "spontaneous validation"
event(cf) <- u
event(cf)
Test for (unique) existence
Description
Group records according to (zero or more) classifying variables. Test for
each group whether at least one (exists) or precisely one
(exists_one) record satisfies a condition.
Usage
exists_any(rule, by = NULL, na.rm = FALSE)
exists_one(rule, by = NULL, na.rm = FALSE)
Arguments
rule |
|
by |
A bare (unquoted) variable name or a list of bare variable names, that will be used to group the data. |
na.rm |
|
Value
A logical vector, with the same number of entries as there
are rows in the entire data under scrutiny. If a test fails, all records in
the group are labeled with FALSE.
See Also
Other cross-record-helpers:
contains_exactly(),
do_by(),
hb(),
hierarchy(),
is_complete(),
is_linear_sequence(),
is_unique()
Examples
# Test whether each household has exactly one 'head of household'
dd <- data.frame(
hhid = c(1, 1, 2, 1, 2, 2, 3 )
, person = c(1, 2, 3, 4, 5, 6, 7 )
, hhrole = c("h","h","m","m","h","m","m")
)
v <- validator(exists_one(hhrole=="h", hhid))
values(confront(dd, v))
# same, but now with missing value in the data
dd <- data.frame(
hhid = c(1, 1, 2, 1, 2, 2, 3 )
, person = c(1, 2, 3, 4, 5, 6, 7 )
, hhrole = c("h",NA,"m","m","h","m","h")
)
values(confront(dd, v))
# same, but now we ignore the missing values
v <- validator(exists_one(hhrole=="h", hhid, na.rm=TRUE))
values(confront(dd, v))
Export to yaml file
Description
Translate an object to yaml format and write to file.
Usage
export_yaml(x, file, ...)
as_yaml(x, ...)
## S4 method for signature 'expressionset'
export_yaml(x, file, ...)
## S4 method for signature 'expressionset'
as_yaml(x, ...)
Arguments
x |
An R object |
file |
A file location or connection (passed to |
... |
Options passed to |
Details
Both validator and indicator objects can be
exported.
Examples
v <- validator(x > 0, y > 0, x + y == z)
txt <- as_yaml(v)
cat(txt)
# NOTE: you can safely run the code below. It is enclosed in 'not run'
# statements to prevent the code from being run at test-time on CRAN
## Not run:
export_yaml(v, file="my_rules.txt")
## End(Not run)
Get expressions
Description
Get expressions
Usage
expr(x, ...)
## S4 method for signature 'rule'
expr(x, ...)
Arguments
x |
Object |
... |
options to be passed to other functions |
Superclass for storing a set of rich expressions.
Description
Superclass for storing a set of rich expressions.
Details
This class is aimed at developers of this package or packages depending on
it, not at users. It is the parent object of both the validator
and the indicator class.
An expressionset is a reference class storing a list of
rules. It contains a number of methods that are not exported
and may change or dissapear without notice. We strongly encourage developers
to use the exported S4 generics to set or extract variables
Exported S4 methods for expressionset
Private S4 methods for expressionset
validating
linear
is_tran_assign
See also
Check whether a field conforms to a regular expression
Description
A convenience wrapper around grepl to make rule sets more readable.
Usage
field_format(x, pattern, type = c("glob", "regex"), ...)
Arguments
x |
Bare (unquoted) name of a variable.
Otherwise a vector of class |
pattern |
|
type |
|
... |
passed to grepl |
See Also
Other format-checkers:
field_length(),
number_format()
Check number of code points
Description
A convenience function testing for field length.
Usage
field_length(x, n = NULL, min = NULL, max = NULL, ...)
Arguments
x |
Bare (unquoted) name of a variable.
Otherwise a vector of class |
n |
Number of code points required. |
min |
Mimimum number of code points |
max |
Maximum number of code points |
... |
passed to |
Value
A [logical] of size length(x).
Details
The number of code points (string length) may depend on current locale
settings or encoding issues, including those caused by inconsistent choices
of UTF normalization.
See Also
Other format-checkers:
field_format(),
number_format()
Examples
df <- data.frame(id = 11001:11003, year = c("2018","2019","2020"), value = 1:3)
rule <- validator(field_length(year, 4), field_length(id, 5))
out <- confront(df, rule)
as.data.frame(out)
Hiridoglu-Berthelot function
Description
A function to measure ‘outlierness’ for skew distributed data with long right tails. The method works by measuring deviation from a reference value, by default the median. Deviation from above is measured as the ratio between observed and refence values. Deviation from below is measured as the inverse: the ratio between reference value and observed values.
Usage
hb(x, ref = stats::median, ...)
Arguments
x |
|
ref |
|
... |
arguments passed to |
Value
\max\{x/ref(x), ref(x)/x\}-1 if ref is a function,
otherwise \max\{x/ref, ref/x\}-1
References
Hidiroglou, M. A., & Berthelot, J. M. (1986). Statistical editing and imputation for periodic business surveys. Survey methodology, 12(1), 73-83.
See Also
Other cross-record-helpers:
contains_exactly(),
do_by(),
exists_any(),
hierarchy(),
is_complete(),
is_linear_sequence(),
is_unique()
Examples
x <- seq(1,20,by=0.1)
plot(x,hb(x), 'l')
Check aggregates defined by a hierarchical code list
Description
Check all aggregates defined by a code hierarchy.
Usage
hierarchy(
values,
labels,
hierarchy,
by = NULL,
tol = 1e-08,
na_value = TRUE,
aggregator = sum,
...
)
Arguments
values |
bare (unquoted) name of a variable that holds values that
must aggregate according to the |
labels |
bare (unquoted) name of variable holding a grouping variable (a code from a hierarchical code list) |
hierarchy |
|
by |
A bare (unquoted) variable or list of variable names that occur in the data under scrutiny. The data will be split into groups according to these variables and the check is performed on each group. |
tol |
|
na_value |
|
aggregator |
|
... |
arguments passed to |
Value
A logical vector with the size of length(values). Every
element involved in an aggregation error is labeled FALSE (aggregate
plus aggregated elements). Elements that are involved in correct
aggregations are set to TRUE, elements that are not involved in
any check get the value na_value (by default: TRUE).
See Also
Other cross-record-helpers:
contains_exactly(),
do_by(),
exists_any(),
hb(),
is_complete(),
is_linear_sequence(),
is_unique()
Examples
# We check some data against the built-in NACE revision 2 classification.
data(nace_rev2)
head(nace_rev2[1:4]) # columns 3 and 4 contain the child-parent relations.
d <- data.frame(
nace = c("01","01.1","01.11","01.12", "01.2")
, volume = c(100 ,70 , 30 ,40 , 25 )
)
# It is possible to perform checks interactively
d$nacecheck <- hierarchy(d$volume, labels = d$nace, hierarchy=nace_rev2[3:4])
# we have that "01.1" == "01.11" + "01.12", but not "01" == "01.1" + "01.2"
print(d)
# Usage as a valiation rule is as follows
rules <- validator(hierarchy(volume, labels = nace, hierarchy=validate::nace_rev_2[3:4]))
confront(d, rules)
# you can also pass a hierarchy as a reference, for example.
rules <- validator(hierarchy(volume, labels = nace, hierarchy=ref$nacecodes))
out <- confront(d, rules, ref=list(nacecodes=nace_rev2[3:4]))
summary(out)
# set a output to NA when a code does not occur in the code list.
d <- data.frame(
nace = c("01","01.1","01.11","01.12", "01.2", "foo")
, volume = c(100 ,70 , 30 ,40 , 25 , 60)
)
d$nacecheck <- hierarchy(d$volume, labels = d$nace, hierarchy=nace_rev2[3:4]
, na_value = NA)
# we have that "01.1" == "01.11" + "01.12", but not "01" == "01.1" + "01.2"
print(d)
Check variable range
Description
Test wether a variable falls within a range.
Usage
in_range(x, min, max, ...)
## Default S3 method:
in_range(x, min, max, strict = FALSE, ...)
## S3 method for class 'character'
in_range(x, min, max, strict = FALSE, format = "auto", ...)
Arguments
x |
A bare (unquoted) variable name. |
min |
lower bound |
max |
upper bound |
... |
arguments passed to other methods |
strict |
|
format |
|
Examples
d <- data.frame(
number = c(3,-2,6)
, time = as.Date(c("2018-02-01", "2018-03-01", "2018-04-01"))
, period = c("2020Q1", "2021Q2", "2020Q3")
)
rules <- validator(
in_range(number, min=-2, max=7, strict=TRUE)
, in_range(time, min=as.Date("2017-01-01"), max=as.Date("2018-12-31"))
, in_range(period, min="2020Q1", max="2020Q4")
)
result <- confront(d, rules)
values(result)
Store results of evaluating indicators
Description
This feature is currently experimental and may change in the future
Details
An indication stores a set of results generated by evaluating
an indicator in the context of data along with some metadata.
Exported S4 methods for indication
Methods exported for objects of class
confrontation
See also
See Also
Other indication-methods:
confront(),
event(),
summary()
Define indicators for data
Description
An indicator maps a data frame, or each record in a data frame to a number. The purpose of this class is to store and apply expressions that define indicators.
Usage
indicator(..., .file, .data)
Arguments
... |
A comma-separated list of indicator definitions |
.file |
(optional) A character vector of file locations |
See Also
Examples
# create an indicator for the number of missing x in data set
I <- indicator(
sum(is.na(.)) # number of missing variables
, sum(is.na(.[c("x","y")])) # number of missing x and y
, mean(is.na(.)) # fraction of missing variables
, sum(x)
, mean(x)
)
dat <- data.frame(x=1:2, y=c(NA,1))
C <- confront(dat, I)
values(C)
Store a set of rich indicator expressions
Description
This feature is currently experimental and may change in future versions
Details
An indicator stores a set of indicators. It is a child class of expressionset and
can be constructed with indicator.
Exported S4 methods for validator
Methods inherited from
expressionset
See also
Test for completeness of records
Description
Utility function to make common tests easier.
Usage
is_complete(...)
all_complete(...)
Arguments
... |
When used in a validation rule: a bare (unquoted) list of variable names. When used directly, a comma-separated list of vectors of equal length. |
Value
For is_complete A logical vector that is FALSE for each record
that has at least one missing value.
For all_unique a single TRUE or FALSE.
See Also
Other cross-record-helpers:
contains_exactly(),
do_by(),
exists_any(),
hb(),
hierarchy(),
is_linear_sequence(),
is_unique()
Examples
d <- data.frame(X = c('a','b',NA,'b'), Y = c(NA,'apple','banana','apple'), Z=1:4)
v <- validator(is_complete(X, Y))
values(confront(d, v))
Check whether a variable represents a linear sequence
Description
A variable X = (x_1, x_2,\ldots, x_n) (n\geq 0) represents a
linear sequence when x_{j+1} - x_j is constant for all
j\geq 1. That is, elements in the series are equidistant and without
gaps.
Usage
is_linear_sequence(x, by = NULL, ...)
## S3 method for class 'numeric'
is_linear_sequence(
x,
by = NULL,
begin = NULL,
end = NULL,
sort = TRUE,
tol = 1e-08,
...
)
## S3 method for class 'Date'
is_linear_sequence(x, by = NULL, begin = NULL, end = NULL, sort = TRUE, ...)
## S3 method for class 'POSIXct'
is_linear_sequence(
x,
by = NULL,
begin = NULL,
end = NULL,
sort = TRUE,
tol = 1e-06,
...
)
## S3 method for class 'character'
is_linear_sequence(
x,
by = NULL,
begin = NULL,
end = NULL,
sort = TRUE,
format = "auto",
...
)
in_linear_sequence(x, ...)
## S3 method for class 'character'
in_linear_sequence(
x,
by = NULL,
begin = NULL,
end = NULL,
sort = TRUE,
format = "auto",
...
)
## S3 method for class 'numeric'
in_linear_sequence(
x,
by = NULL,
begin = NULL,
end = NULL,
sort = TRUE,
tol = 1e-08,
...
)
## S3 method for class 'Date'
in_linear_sequence(x, by = NULL, begin = NULL, end = NULL, sort = TRUE, ...)
## S3 method for class 'POSIXct'
in_linear_sequence(
x,
by = NULL,
begin = NULL,
end = NULL,
sort = TRUE,
tol = 1e-06,
...
)
Arguments
x |
An R vector. |
by |
bare (unquoted) variable name or a list of unquoted variable names,
used to split |
... |
Arguments passed to other methods. |
begin |
Optionally, a value that should equal |
end |
Optionally, a value that should equal |
sort |
|
tol |
numerical tolerance for gaps. |
format |
|
Details
Presence of a missing value (NA) in x will result in NA,
except when length(x) <= 2 and start and end are
NULL. Any sequence of length \leq 2 is a linear sequence.
Value
For is_linear_sequence: a single TRUE or FALSE,
equal to all(in_linear_sequence).
For in_linear_sequence: a logical vector with the same length as x.
See Also
Other cross-record-helpers:
contains_exactly(),
do_by(),
exists_any(),
hb(),
hierarchy(),
is_complete(),
is_unique()
Examples
is_linear_sequence(1:5) # TRUE
is_linear_sequence(c(1,3,5,4,2)) # FALSE
is_linear_sequence(c(1,3,5,4,2), sort=TRUE) # TRUE
is_linear_sequence(NA_integer_) # TRUE
is_linear_sequence(NA_integer_, begin=4) # FALSE
is_linear_sequence(c(1, NA, 3)) # FALSE
d <- data.frame(
number = c(pi, exp(1), 7)
, date = as.Date(c("2015-12-17","2015-12-19","2015-12-21"))
, time = as.POSIXct(c("2015-12-17","2015-12-19","2015-12-20"))
)
rules <- validator(
is_linear_sequence(number) # fails
, is_linear_sequence(date) # passes
, is_linear_sequence(time) # fails
)
summary(confront(d,rules))
## check groupwise data
dat <- data.frame(
time = c(2012, 2013, 2012, 2013, 2015)
, type = c("hi", "hi", "ha", "ha", "ha")
)
rule <- validator(in_linear_sequence(time, by=type))
values(confront(dat, rule)) ## 2xT, 3xF
rule <- validator(in_linear_sequence(time, type))
values( confront(dat, rule) )
Test for uniquenes of records
Description
Test for uniqueness of columns or combinations of columns.
Usage
is_unique(...)
all_unique(...)
n_unique(...)
Arguments
... |
When used in a validation rule: a bare (unquoted) list of variable names. When used directly, a comma-separated list of vectors of equal length. |
Value
For is_unique A logical vector that is FALSE for each record
that has a duplicate.
For all_unique a single TRUE or FALSE.
For number_unique a single number representing the number
of unique values or value combinations in the arguments.
See Also
Other cross-record-helpers:
contains_exactly(),
do_by(),
exists_any(),
hb(),
hierarchy(),
is_complete(),
is_linear_sequence()
Examples
d <- data.frame(X = c('a','b','c','b'), Y = c('banana','apple','banana','apple'), Z=1:4)
v <- validator(is_unique(X, Y))
values(confront(d, v))
# example with groupwise test
df <- data.frame(x=c(rep("a",3), rep("b",3)),y=c(1,1,2,1:3))
v <- validator(is_unique(y, by=x))
values(confront(d,v))
Get key set stored with a confrontation
Description
Get key set stored with a confrontation
Usage
keyset(x)
## S4 method for signature 'confrontation'
keyset(x)
Arguments
x |
an object of class |
Value
If a confrontation is created with the key= option
set, this function returns the key set, otherwise NULL
See Also
Other confrontation-methods:
[,expressionset-method,
as.data.frame,confrontation-method,
confront(),
confrontation-class,
errors(),
event(),
length,expressionset-method,
values()
Rule label
Description
A short (typically two or three word) description of a rule.
Usage
label(x, ...)
label(x) <- value
## S4 method for signature 'rule'
label(x, ...)
## S4 replacement method for signature 'rule,character'
label(x) <- value
## S4 method for signature 'expressionset'
label(x, ...)
## S4 replacement method for signature 'expressionset,character'
label(x) <- value
Arguments
x |
and R object |
... |
Arguments to be passed to other methods |
value |
Value to set |
Value
A character vector.
See Also
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
description(),
meta(),
names<-,rule,character-method,
origin(),
plot,validator-method,
summary(),
variables(),
voptions()
Examples
# retrieve properties
v <- validator(turnover > 0, staff.costs>0)
# number of rules in v:
length(v)
# per-rule
created(v)
origin(v)
names(v)
# set properties
names(v)[1] <- "p1"
label(v)[1] <- "turnover positive"
description(v)[1] <- "
According to the official definition,
only positive values can be considered
valid turnovers.
"
# short description is also printed:
v
# print all info for first rule
v[[1]]
# retrieve properties
v <- validator(turnover > 0, staff.costs>0)
# number of rules in v:
length(v)
# per-rule
created(v)
origin(v)
names(v)
# set properties
names(v)[1] <- "p1"
label(v)[1] <- "turnover positive"
description(v)[1] <- "
According to the official definition,
only positive values can be considered
valid turnovers.
"
# short description is also printed:
v
# print all info for first rule
v[[1]]
Logging object to use with the lumberjack package
Description
Logging object to use with the lumberjack package
Format
A reference class object
Methods
add(meta, input, output)Add logging info based on in- and output
dump(file = NULL, verbose = TRUE, ...)Dump logging info to csv file. All arguments in '...' except row.names are passed to 'write.csv'
initialize(..., verbose = TRUE, label = "")Create object. Optionally toggle verbosity.
log_data()Return logged data as a data.frame
Details
This obeject can used with the function composition ('pipe') operator of the
lumberjack package. The logging is based on
validate's cells function. The output is written to a
csv file wich contains the following columns.
step | integer | Step number |
time | POSIXct | Timestamp |
expr | character | Expression used on data |
cells | integer | Total nr of cells in dataset |
available | integer | Nr of non-NA cells |
missing | integer | Nr of empty (NA) cells |
still_available | integer | Nr of cells still available after expr |
unadapted | integer | Nr of cells still available and unaltered |
unadapted | integer | Nr of cells still available and altered |
imputed | integer | Nr of cells not missing anymore |
Note
This logger is suited only for operations that do not change the dimensions of the dataset.
See Also
Other loggers:
lbj_rules-class
Logging object to use with the lumberjack package
Description
Logging object to use with the lumberjack package
Methods
dump(file = NULL, ...)Dump logging info to csv file. All arguments in '...' except row.names are passed to 'write.csv'
initialize(rules, verbose = TRUE, label = "")Create object. Optionally toggle verbosity.
log_data()Return logged data as a data.frame
plot()plot rule comparisons
See Also
Other loggers:
lbj_cells-class
Determine the number of elements in an object.
Description
Determine the number of elements in an object.
Usage
## S4 method for signature 'expressionset'
length(x)
## S4 method for signature 'confrontation'
length(x)
Arguments
x |
An R object |
See Also
Other confrontation-methods:
[,expressionset-method,
as.data.frame,confrontation-method,
confront(),
confrontation-class,
errors(),
event(),
keyset(),
values()
Create matching subsets of a sequence of data
Description
Create matching subsets of a sequence of data
Usage
match_cells(..., .list = NULL, id = NULL)
Arguments
... |
A sequence of |
.list |
A list of |
id |
Names or indices of columns to use as index. |
Value
A list of data.frames, subsetted and sorted so that all cells correspond.
See Also
Other comparing:
as.data.frame,cellComparison-method,
as.data.frame,validatorComparison-method,
barplot,cellComparison-method,
barplot,validatorComparison-method,
cells(),
compare(),
plot,cellComparison-method,
plot,validatorComparison-method
Get or set rule metadata
Description
Rule metadata are key-value pairs where the value is a simple (atomic) string or number.
Usage
meta(x, ...)
meta(x, name) <- value
## S4 method for signature 'rule'
meta(x, ...)
## S4 replacement method for signature 'rule,character'
meta(x, name) <- value
## S4 method for signature 'expressionset'
meta(x, simplify = TRUE, ...)
## S4 replacement method for signature 'expressionset,character'
meta(x, name) <- value
Arguments
x |
an R object |
... |
Arguments to be passed to other methods |
name |
|
value |
Value to set |
simplify |
Gather all metadata into a dataframe? |
See Also
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
description(),
label(),
names<-,rule,character-method,
origin(),
plot,validator-method,
summary(),
variables(),
voptions()
Examples
v <- validator(x > 0, y > 0)
# metadata is recycled over rules
meta(v,"foo") <- "bar"
# assign metadata to a selection of rules
meta(v[1],"fu") <- 2
# retrieve metadata as data.frame
meta(v)
# retrieve metadata as list
meta(v,simplify=TRUE)
NACE classification code table
Description
Statistical Classification of Economic Activities.
Order
[integer]Level
[integer]NACE levelCode
[character]NACE codeParent
[character]parent code of"Code"Description
[character]This_item_includes
[character]This_item_also_includes
[character]Rulings
[character]This_item_excludes
[character]Reference_to_ISIC_Rev._4
[character]
Format
A csv file, one NACE code per row.
See Also
Other datasets:
retailers,
samplonomy
Extract or set names
Description
Extract or set names
When setting names, values are recycled and made unique with
make.names
Get names from confrontation object
Usage
## S4 replacement method for signature 'rule,character'
names(x) <- value
## S4 method for signature 'expressionset'
names(x)
## S4 replacement method for signature 'expressionset,character'
names(x) <- value
## S4 method for signature 'confrontation'
names(x)
Arguments
x |
An R object |
value |
Value to set |
Value
A character vector
See Also
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
description(),
label(),
meta(),
origin(),
plot,validator-method,
summary(),
variables(),
voptions()
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
compare(),
confront(),
event(),
plot,validation-method,
sort,validation-method,
summary(),
validation-class,
values()
Examples
# retrieve properties
v <- validator(turnover > 0, staff.costs>0)
# number of rules in v:
length(v)
# per-rule
created(v)
origin(v)
names(v)
# set properties
names(v)[1] <- "p1"
label(v)[1] <- "turnover positive"
description(v)[1] <- "
According to the official definition,
only positive values can be considered
valid turnovers.
"
# short description is also printed:
v
# print all info for first rule
v[[1]]
# retrieve properties
v <- validator(turnover > 0, staff.costs>0)
# number of rules in v:
length(v)
# per-rule
created(v)
origin(v)
names(v)
# set properties
names(v)[1] <- "p1"
label(v)[1] <- "turnover positive"
description(v)[1] <- "
According to the official definition,
only positive values can be considered
valid turnovers.
"
# short description is also printed:
v
# print all info for first rule
v[[1]]
Check the layouts of numbers.
Description
Convenience function to check layout of numbers stored as a character vector.
Usage
number_format(x, format = NULL, min_dig = NULL, max_dig = NULL, dec = ".")
Arguments
x |
|
format |
|
min_dig |
|
max_dig |
|
dec |
|
Details
If format is specified, then min_dig, max_dig and dec
are ignored.
Numerical formats can be specified as a sequence of characters. There are a few special characters:
dStands for digit.*(digit globbing) zero or more digits
Here are some examples.
"d.dd" | One digit, a decimal point followed by two digits. |
"d.ddddddddEdd" | Scientific notation with eight digits behind the decimal point. |
"0.ddddddddEdd" | Same, but starting with a zero. |
"d,dd*" | one digit before the comma and at least two behind it. |
See Also
Other format-checkers:
field_format(),
field_length()
Examples
df <- data.frame(number = c("12.34","0.23E55","0.98765E12"))
rules <- validator(
number_format(number, format="dd.dd")
, number_format(number, "0.ddEdd")
, number_format(number, "0.*Edd")
)
out <- confront(df, rules)
values(out)
# a few examples, without 'validator'
number_format("12.345", min_dig=2) # TRUE
number_format("12.345", min_dig=4) # FALSE
number_format("12.345", max_dig=2) # FALSE
number_format("12.345", max_dig=5) # TRUE
number_format("12,345", min_dig=2, max_dig=3, dec=",") # TRUE
Origin of rules
Description
A slot to store where the rule originated, e.g. a filename
or "command-line" for interactively defined rules.
Usage
origin(x, ...)
origin(x) <- value
## S4 method for signature 'rule'
origin(x, ...)
## S4 replacement method for signature 'rule,character'
origin(x) <- value
## S4 method for signature 'expressionset'
origin(x, ...)
## S4 replacement method for signature 'expressionset,character'
origin(x) <- value
Arguments
x |
and R object |
... |
Arguments to be passed to other methods |
value |
Value to set |
Value
A character vector.
See Also
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
description(),
label(),
meta(),
names<-,rule,character-method,
plot,validator-method,
summary(),
variables(),
voptions()
Examples
# retrieve properties
v <- validator(turnover > 0, staff.costs>0)
# number of rules in v:
length(v)
# per-rule
created(v)
origin(v)
names(v)
# set properties
names(v)[1] <- "p1"
label(v)[1] <- "turnover positive"
description(v)[1] <- "
According to the official definition,
only positive values can be considered
valid turnovers.
"
# short description is also printed:
v
# print all info for first rule
v[[1]]
# retrieve properties
v <- validator(turnover > 0, staff.costs>0)
# number of rules in v:
length(v)
# per-rule
created(v)
origin(v)
names(v)
# set properties
names(v)[1] <- "p1"
label(v)[1] <- "turnover positive"
description(v)[1] <- "
According to the official definition,
only positive values can be considered
valid turnovers.
"
# short description is also printed:
v
# print all info for first rule
v[[1]]
Test whether details combine to a chosen aggregate
Description
Data in 'long' format often contain records representing totals (or other aggregates) as well as records that contain details that add up to the total. This function facilitates checking the part-whole relation in such cases.
Usage
part_whole_relation(
values,
labels,
whole,
part = NULL,
aggregator = sum,
tol = 1e-08,
by = NULL,
...
)
Arguments
values |
A bare (unquoted) variable name holding the values to aggregate |
labels |
A bare (unquoted) variable name holding the labels indicating whether a value is an aggregate or a detail. |
whole |
|
part |
|
aggregator |
|
tol |
|
by |
Name of variable, or |
... |
Extra arguments passed to aggregator (for example |
Value
A logical vector of size length(value).
Examples
df <- data.frame(
id = 10011:10020
, period = rep(c("2018Q1", "2018Q2", "2018Q3", "2018Q4","2018"),2)
, direction = c(rep("import",5), rep("export", 5))
, value = c(1,2,3,4,10, 3,3,3,3,13)
)
## use 'rx' to interpret 'whole' as a regular expression.
rules <- validator(
part_whole_relation(value, period, whole=rx("^\\d{4}$")
, by=direction)
)
out <- confront(df, rules, key="id")
as.data.frame(out)
Line graph of a cellComparison object.
Description
Versions of a data set can be compared cell by cell
using cells. The result is a cellComparison
object. This method creates a line-graph, thus suggesting an
that an ordered sequence of data sets have been compared.
See also barplot,cellComparison-method for an
unordered version.
Usage
## S4 method for signature 'cellComparison'
plot(x, xlab = "", ylab = "", las = 2, cex.axis = 0.8, cex.legend = 0.8, ...)
Arguments
x |
a |
xlab |
[ |
ylab |
[ |
las |
[ |
cex.axis |
[ |
cex.legend |
[ |
... |
Graphical parameters, passed to |
See Also
Other comparing:
as.data.frame,cellComparison-method,
as.data.frame,validatorComparison-method,
barplot,cellComparison-method,
barplot,validatorComparison-method,
cells(),
compare(),
match_cells(),
plot,validatorComparison-method
Plot validation results
Description
Creates a barplot of validation result. For each validation rule, a stacked bar is plotted with percentages of failing, passing, and missing results.
Usage
## S4 method for signature 'validation'
plot(
x,
y,
fill = c("#FE2712", "#66B032", "#dddddd"),
col = fill,
rulenames = names(x),
labels = c("Fails", "Passing", "Missing", "Total"),
title = NULL,
xlab = NULL,
...
)
Arguments
x |
a confrontation object. |
y |
not used |
fill |
|
col |
Edge colors for the bars. |
rulenames |
|
labels |
|
title |
|
xlab |
|
... |
not used |
Details
The plot function tries to be smart about placing labels on the y axis. When the number of bars becomes too large, no y axis annotation will be shown and the bars will become space-filling.
See Also
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
compare(),
confront(),
event(),
names<-,rule,character-method,
sort,validation-method,
summary(),
validation-class,
values()
Examples
rules <- validator( r1 = staff.costs < total.costs
, r2 = turnover + other.rev == total.rev
, r3 = other.rev > 0
, r4 = total.rev > 0
, r5 = nace %in% c("A", "B")
)
plot(rules, cex=0.8, show_legend=TRUE)
data(retailers)
cf <- confront(retailers, rules)
plot(cf, main="Retailers check")
Plot a validator object
Description
The matrix of variables x rules is plotted, in which rules that are
recognized as linear (in)equations are differently colored.
The augmented matrix is returned, but can also be calculated using
variables(x, as="matrix").
Usage
## S4 method for signature 'validator'
plot(
x,
y,
use_blocks = TRUE,
col = c("#b2df8a", "#a6cee3"),
cex = 1,
show_legend = TRUE,
...
)
Arguments
x |
validator object with rules |
y |
not used |
use_blocks |
|
col |
|
cex |
size of the variables plotted. |
show_legend |
should a legend explaining the colors be drawn? |
... |
passed to image |
Value
(invisible) the matrix
See Also
Other validator-methods:
+,validator,validator-method,
validator
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
description(),
label(),
meta(),
names<-,rule,character-method,
origin(),
summary(),
variables(),
voptions()
Examples
rules <- validator( r1 = staff.costs < total.costs
, r2 = turnover + other.rev == total.rev
, r3 = other.rev > 0
, r4 = total.rev > 0
, r5 = nace %in% c("A", "B")
)
plot(rules, cex=0.8, show_legend=TRUE)
data(retailers)
cf <- confront(retailers, rules)
plot(cf, main="Retailers check")
Line graph of validatorComparison object
Description
The performance of versions of a data set with regard to rule-based quality
requirements can be compared using using compare. The result is a
validatorComparison object. This method creates a line-graph, thus
suggesting an that an ordered sequence of data sets have been compared. See
also barplot,validatorComparison-method for an unordered version.
Usage
## S4 method for signature 'validatorComparison'
plot(x, xlab = "", ylab = "", las = 2, cex.axis = 0.8, cex.legend = 0.8, ...)
Arguments
x |
Object of class |
xlab |
[ |
ylab |
[ |
las |
[ |
cex.axis |
[ |
cex.legend |
[ |
... |
Graphical parameters, passed to |
See Also
Other comparing:
as.data.frame,cellComparison-method,
as.data.frame,validatorComparison-method,
barplot,cellComparison-method,
barplot,validatorComparison-method,
cells(),
compare(),
match_cells(),
plot,cellComparison-method
data on Dutch supermarkets
Description
Anonymized and distorted data on revenue and cost structure for
60 retailers. Currency is in thousands of Euros. There are two
data sets. The SBS2000 dataset is equal to the retailers
data set except that it has a record identifier (called id) column.
id: A unique identifier (only in SBS2000)
size: Size class (0=undetermined)
incl.prob: Probability of inclusion in the sample
staff: Number of staff
turnover: Amount of turnover
other.rev: Amount of other revenue
total.rev: Total revenue
staff.costs: Costs assiciated to staff
total.costs: Total costs made
profit: Amount of profit
vat: Turnover reported for Value Added Tax
Format
A csv file, one retailer per row.
See Also
Other datasets:
nace_rev2,
samplonomy
A rich expression
Description
A rich expression
Details
Technically, rule is a call object endowed with extra
attributes such as a name, a label and a description description, creation
time and a reference to its origin. Rule objects are not for direct use by
users of the package, but may be of interest for developers of this package,
or packages depending on it.
Exported S4 methods for rule
show
Private S4 methods for rule
validating
linear
expr
is_tran_assign
See also
Run a file with confrontations. Capture results
Description
A validation script is a regular R script, intersperced with confront
or check_that statements. This function will run the script file
and capture all output from calls to these functions.
Usage
run_validation_file(file, verbose = TRUE)
run_validation_dir(dir = "./", pattern = "^validate.+[rR]", verbose = TRUE)
## S3 method for class 'validations'
print(x, ...)
## S3 method for class 'validations'
summary(object, ...)
Arguments
file |
|
verbose |
|
dir |
|
pattern |
|
x |
An R object |
... |
Unused |
object |
An R object |
Value
run_validation_file: An object of class validations. This is
a list of objects of class validation.
run_validation_dir: An object of class validations. This is
a list of objects of class validation.
print: NULL, invisibly.
summary: A data frame similar to the data frame returned
when summarizing a validation object. There are extra columns listing
each call, file and first and last line where the code occurred.
Label objects for interpretation as pattern
Description
Label objects (typically strings or data frames containing keys combinations) to be interpreted as regular expression or globbing pattern.
Usage
rx(x)
glob(x)
Arguments
x |
Object to label as regular expression ( |
Economic data on Samplonia
Description
Simulated economic time series representing GDP, Import, Export and Balance of Trade (BOT) of Samplonia. Samplonia is a fictional Island invented by Jelke Bethelehem (2009). The country has 10 000 inhabitants. It consists of two provinces: Agria and Induston. Agria is a rural province consisting of the mostly fruit and vegetable producing district of Wheaton and the mostly cattle producing Greenham. Induston has four districts. Two districts with heavy industry named Smokeley and Mudwater. Newbay is a young, developing district while Crowdon is where the rich Samplonians retire. The current data set contains several time series from Samplonia's national accounts system in long format.
There are annual and quarterly time series on GDP, Import, Export and Balance of Trade, for Samplonia as a whole, for each province and each district. BOT is defined as Export-Import for each region and period; quarterly figures are expected to add up to annual figures for each region and measure, and subregions are expected to add up to their super-regions.
region: Region (Samplonia, one if its 2 provinces, or one of its 6 districts)
freq: Frequency of the time series
period: Period (year or quarter)
measure: The economic variable (gdp, import, export, balance)
value: The value
The data set has been endowed with the following errors.
For Agria, the 2015 GDP record is not present.
For Induston, the 2018Q3 export value is missing (
NA)For Induston, there are two different values for the 2018Q2 Export
For Crowdon, the 2015Q1 balance value is missing (
NA).For Wheaton, the 2019Q2 import is missing (
NA).
Format
An RData file.
References
J. Bethlehem (2009), Applied Survey Methods: A Statistical Perspective. John Wiley & Sons, Hoboken, NJ.
See Also
Other datasets:
nace_rev2,
retailers
Select records (not) satisfying rules
Description
Apply validation rules or validation results to a data set and select only those that satisfy all or violate at least one rule.
Usage
satisfying(x, y, include_missing = FALSE, ...)
violating(x, y, include_missing = FALSE, ...)
## Default S3 method:
violating(x, y, include_missing = FALSE, ...)
lacking(x, y, ...)
Arguments
x |
A |
y |
a |
include_missing |
Toggle: also select records that have |
... |
options passed to |
Value
For satisfying, the records in x satisfying all rules or
validation outcomes in y. For violating the records in
x violating at least one of the rules or validation outcomes
in y
Note
An error is thrown if the rules or validation results in y can not be
interpreted record-by record (e.g. when one of the rules is of the form
mean(foo)>0).
Examples
rules <- validator(speed >= 12, dist < 100)
satisfying(cars, rules)
violating(cars, rules)
out <- confront(cars, rules)
summary(out)
satisfying(cars, out)
violating(cars, out)
Get code list from an SDMX REST API endpoint.
Description
sdmx_codelist constructs an URL for rsdmx::readSDMX and
extracts the code IDs. Code lists are downloaded once and cached for the
duration of the R session.
estat_codelist gets a code list from the REST API provided at
ec.europa.eu/tools/cspa_services_global/sdmxregistry. It is a
convenience wrapper that calls sdmx_codelist.
global_codelist gets a code list from the REST API provided at
https://registry.sdmx.org/webservice/data.html. It is a convenience
wrapper that calls sdmx_codelist.
Usage
sdmx_codelist(
endpoint,
agency_id,
resource_id,
version = "latest",
what = c("id", "all")
)
estat_codelist(resource_id, agency_id = "ESTAT", version = "latest")
global_codelist(resource_id, agency_id = "SDMX", version = "latest")
Arguments
endpoint |
|
agency_id |
|
resource_id |
|
version |
|
what |
|
See Also
Other sdmx:
sdmx_endpoint(),
validator_from_dsd()
Other sdmx:
sdmx_endpoint(),
validator_from_dsd()
Examples
# here we download the CL_ACTIVITY codelist from the ESTAT registry.
## Not run:
codelist <- sdmx_codelist(
endpoint = "https://registry.sdmx.org/ws/public/sdmxapi/rest/"
, agency_id = "ESTAT"
, resource_id = "CL_ACTIVITY"
## End(Not run)
## Not run:
estat_codelist("CL_ACTIVITY")
## End(Not run)
## Not run:
global_codelist("CL_AGE") )
global_codelist("CL_CONF_STATUS")
global_codelist("CL_SEX")
## End(Not run)
# An example of using SDMX information, downloaded from the SDMX global
# registry
## Not run:
# economic data from the country of Samplonia
data(samplonomy)
head(samplonomy)
rules <- validator(
, freq %in% global_codelist("CL_FREQ")
, value >= 0
)
cf <- confront(samplonomy, rules)
summary(cf)
## End(Not run)
Get URL for known SDMX registry endpoints
Description
Convenience function storing URLs for SDMX endpoints.
Usage
sdmx_endpoint(registry = NULL)
Arguments
registry |
|
See Also
Other sdmx:
sdmx_codelist(),
validator_from_dsd()
Examples
sdmx_endpoint()
sdmx_endpoint("ESTAT")
sdmx_endpoint("global")
Aggregate and sort the results of a validation.
Description
Aggregate and sort the results of a validation.
Usage
## S4 method for signature 'validation'
sort(x, decreasing = FALSE, by = c("rule", "record"), drop = TRUE, ...)
Arguments
x |
An object of class |
decreasing |
Sort by decreasing number of passes? |
by |
Report on violations per rule (default) or per record? |
drop |
drop list attribute if the result has a single argument. |
... |
Arguments to be passed to or from other methods. |
Value
A data.frame with the following columns.
| keys | If confront was called with key= |
npass | Number of items passed |
nfail | Number of items failing |
nNA | Number of items resulting in NA |
rel.pass | Relative number of items passed |
rel.fail | Relative number of items failing |
rel.NA | Relative number of items resulting in NA
|
If by='rule' the relative numbers are computed with respect to the number
of records for which the rule was evaluated. If by='record' the relative numbers
are computed with respect to the number of rules the record was tested agains. By default
the most failed validations and records with the most fails are on the top.
When by='record' and not all validation results have the same dimension structure,
a list of data.frames is returned.
See Also
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
compare(),
confront(),
event(),
names<-,rule,character-method,
plot,validation-method,
summary(),
validation-class,
values()
Examples
data(retailers)
retailers$id <- paste0("ret",1:nrow(retailers))
v <- validator(
staff.costs/staff < 25
, turnover + other.rev==total.rev)
cf <- confront(retailers,v,key="id")
a <- aggregate(cf,by='record')
head(a)
# or, get a sorted result:
s <- sort(cf, by='record')
head(s)
Create a summary
Description
Create a summary
Usage
summary(object, ...)
## S4 method for signature 'expressionset'
summary(object, ...)
## S4 method for signature 'indication'
summary(object, ...)
## S4 method for signature 'validation'
summary(object, ...)
Arguments
object |
An R object |
... |
Currently unused |
Value
A data.frame with the information mentioned below is returned.
Validator and indicator objects
For these objects, the ruleset is split into subsets (blocks) that are disjunct in the sense that they do not share any variables. For each block the number of variables, the number of rules and the number of rules that are linear are reported.
Indication
Some basic information per evaluated indicator is reported: the number
of items to which the indicator was applied, the output class,
some statistics (min, max, mean , number of NA)
and wether an exception occurred (warnings or errors). The evaluated
expression is reported as well.
Validation
Some basic information per evaluated validation rule is reported: the number of
items to which the rule was applied, the output class, some statistics
(passes, fails, number of NA) and wether an exception occurred (warnings or
errors). The evaluated expression is reported as well.
See Also
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
description(),
label(),
meta(),
names<-,rule,character-method,
origin(),
plot,validator-method,
variables(),
voptions()
Other indication-methods:
confront(),
event(),
indication-class
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
compare(),
confront(),
event(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
validation-class,
values()
Examples
data(retailers)
v <- validator(staff > 0, staff.costs/staff < 20, turnover+other.revenue == total.revenue)
summary(v)
cf <- confront(retailers,v)
summary(cf)
Syntax to define validation or indicator rules
Description
A concise overview of the validate syntax.
Basic syntax
The basic rule is that an R-statement that evaluates to a logical is a
validating statement. This is established by static code inspection when
validator reads a (set of) user-defined validation rule(s).
Comparisons
All basic comparisons, including >, >=, ==, !=, <=, <, %in%
are validating statements. When executing a validating statement, the
%in% operator is replaced with %vin%.
Logical operations
Unary logical operators '!', all() and any define
validating statements. Binary logical operations including &, &&, |,
||, are validating when P and Q in e.g. P & Q are
validating. (note that the short-circuits && and & onnly return
the first logical value, in cases where for P && Q, P and/or
Q are vectors. Binary logical implication P\Rightarrow Q (P
implies Q) is implemented as if ( P ) Q. The latter is interpreted as
!(P) | Q.
Type checking
Any function starting with is. (e.g. is.numeric) is a
validating expression.
Text search
grepl, nzchar, startsWith, and endsWith are validating
expressions.
Functional dependencies
Armstrong's functional dependencies, of the form A + B \to C + D are
represented using the ~, e.g. A + B ~ C + D. For example
postcode ~ city means, that when two records have the same value for
postcode, they must have the same value for city.
Reference the dataset as a whole
Metadata such as numer of rows, columns, column names and so on can be
tested by referencing the whole data set with the '.'. For example,
the rule nrow(.) == 15 checks whether there are 15 rows in the
dataset at hand.
Uniqueness, completeness
These can be tested in principle with the 'dot' syntax. However, there are
some convenience functions: is_complete, all_complete
is_unique, all_unique.
Local, transient assignment
The operator ':=' can be used to set up local variables (during, for
example, validation) to save time (the rhs of an assignment is computed only
once) or to make your validation code more maintainable. Assignments work more
or less like common R assignments: they are only valid for statements coming
after the assignment and they may be overwritten. The result of computing the
rhs is not part of a confrontation with data.
Groups
Often the same constraints/rules are valid for groups of variables.
validate allows for compact notation. Variable groups can be used
in-statement or by defining them with the := operator.
validator( var_group(a,b) > 0 )
is equivalent to
validator(G := var_group(a,b), G > 0)
is equivalent to
validator(a>0,b>0).
Using two groups results in the cartesian product of checks. So the statement
validator( f=var_group(c,d), g=var_group(a,b), g > f)
is equivalent to
validator(a > c, b > c, a > d, b > d)
File parsing
Please see the cookbook on how to read rules from and write rules to file:
vignette("cookbook",package="validate")
Store results of evaluating validating expressions
Description
Store results of evaluating validating expressions
Details
A object of class validation stores a set of results generated by
evaluating an validator in the context of data along with some
metadata.
See Also
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
compare(),
confront(),
event(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
summary(),
values()
Define validation rules for data
Description
Define validation rules for data
Usage
validator(..., .file, .data)
Arguments
... |
A comma-separated list of validating expressions |
.file |
(optional) A character vector of file locations (see also the
section on file parsing in the |
.data |
(optional) A |
Value
An object of class validator (see validator-class).
Validating expressions
Each validating expression should evaluate to a logical. Allowed syntax of
the expression is described in syntax.
See Also
Other validator-methods:
+,validator,validator-method,
plot,validator-method
Examples
v <- validator(
height>0
,weight>0
,height < 1.5*mean(height)
)
cf <- confront(women, v)
summary(cf)
Store a set of rich validating rules.
Description
Store a set of rich validating rules.
Details
A validator stores a set of validatin rules. It is a child class of
expressionset and
can be constructed with validator. validator contains
an extra slot "language" stating the language in which the validation
rule is expressed. The default, and currently only supported language is
the validate language implemented by this package.
Exported S4 methods for validator
Methods inherited from
expressionset
See also
Extract a rule set from an SDMX DSD file
Description
Data Structure Definitions contain references to code lists. This function extracts those references and generates rules that check data against code lists in an SDMX registry.
Usage
validator_from_dsd(endpoint, agency_id, resource_id, version = "latest")
Arguments
endpoint |
|
agency_id |
|
resource_id |
|
version |
|
Value
An object of class validator.
See Also
Other sdmx:
sdmx_codelist(),
sdmx_endpoint()
Get values from object
Description
Get values from object
Usage
values(x, ...)
## S4 method for signature 'confrontation'
values(x, ...)
## S4 method for signature 'validation'
values(x, simplify = TRUE, drop = TRUE, ...)
## S4 method for signature 'indication'
values(x, simplify = TRUE, drop = TRUE, ...)
Arguments
x |
an R object |
... |
Arguments to pass to or from other methods |
simplify |
Combine results with similar dimension structure into arrays? |
drop |
if a single vector or array results, drop 'list' attribute? |
See Also
Other confrontation-methods:
[,expressionset-method,
as.data.frame,confrontation-method,
confront(),
confrontation-class,
errors(),
event(),
keyset(),
length,expressionset-method
Other validation-methods:
aggregate,validation-method,
all,validation-method,
any,validation-method,
barplot,validation-method,
check_that(),
compare(),
confront(),
event(),
names<-,rule,character-method,
plot,validation-method,
sort,validation-method,
summary(),
validation-class
Get variable names
Description
Generic function that extracts names of variables ocurring in R objects.
Usage
variables(x, ...)
## S4 method for signature 'rule'
variables(x, ...)
## S4 method for signature 'list'
variables(x, ...)
## S4 method for signature 'data.frame'
variables(x, ...)
## S4 method for signature 'environment'
variables(x, ...)
## S4 method for signature 'expressionset'
variables(x, as = c("vector", "matrix", "list"), dummy = FALSE, ...)
Arguments
x |
An R object |
... |
Arguments to be passed to other methods. |
as |
how to return variables:
|
dummy |
Also retrieve transient variables set with the |
Methods (by class)
-
variables(rule): Retrieve unique variable names -
variables(list): Alias tonames.list -
variables(data.frame): Alias tonames.data.frame -
variables(environment): Alias tols -
variables(expressionset): Variables occuring inxeither as a single list, or per rule.
See Also
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
description(),
label(),
meta(),
names<-,rule,character-method,
origin(),
plot,validator-method,
summary(),
voptions()
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
description(),
label(),
meta(),
names<-,rule,character-method,
origin(),
plot,validator-method,
summary(),
voptions()
Examples
v <- validator(
root = y := sqrt(x)
, average = mean(x) > 3
, sum = x + y == z
)
variables(v)
variables(v,dummy=TRUE)
variables(v,matrix=TRUE)
variables(v,matrix=TRUE,dummy=TRUE)
Set or get options globally or per object.
Description
There are three ways to specify options for this package.
Globally. Setting
voptions(option1=value1,option2=value2,...)sets global options.Per object. Setting
voptions(x=<object>, option1=value1,...), causes all relevant functions that use that object (e.g.confront) to use those local settings.At execution time. Relevant functions (e.g.
confront) take optional arguments allowing one to define options to be used during the current function call
Usage
voptions(x = NULL, ...)
## S4 method for signature 'ANY'
voptions(x = NULL, ...)
validate_options(...)
reset(x = NULL)
## S4 method for signature 'ANY'
reset(x = NULL)
## S4 method for signature 'expressionset'
voptions(x = NULL, ...)
## S4 method for signature 'expressionset'
reset(x = NULL)
Arguments
x |
(optional) an object inheriting from |
... |
Name of an option (character) to retrieve options or |
Value
When requesting option settings: a list. When setting options,
the whole options list is returned silently.
Options for the validate package
Currently the following options are supported.
na.value(NA,TRUE,FALSE;NA) Value to return when a validating statement results inNA.raise("none","error","all";"none") Control if theconfrontmethods catch or raise exceptions. The 'all' setting is useful when debugging validation scripts.lin.eq.eps('numeric'; 1e-8) The precision used when evaluating linear equalities. To be used to control for machine rounding."reset"Reset to factory settings.
See Also
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
description(),
label(),
meta(),
names<-,rule,character-method,
origin(),
plot,validator-method,
summary(),
variables()
Other expressionset-methods:
as.data.frame(),
as.data.frame,expressionset-method,
created(),
description(),
label(),
meta(),
names<-,rule,character-method,
origin(),
plot,validator-method,
summary(),
variables()
Examples
# set an option, local to a validator object:
v <- validator(x + y > z)
voptions(v,raise='all')
# check that local option was set:
voptions(v,'raise')
# check that global options have not changed:
voptions('raise')